Tutorials
ipyrad command line assembly
This is the full tutorial for the command line interface (CLI) for ipyrad. In this tutorial we’ll walk through the entire assembly, from raw data to output files for downstream analysis. This is meant as a broad introduction to familiarize users with the general workflow, and some of the parameters and terminology. We will use and empirical dataset of paired-end cheetah ddRAD data as an example in this tutorial. Of course, you can replicate the steps described here with your own data, or any other RADseq dataset.
If you are new to RADseq analyses, this tutorial will provide a simple overview of how to execute ipyrad, what the data files look like, how to check that your analysis is working, and what the final output formats will be. We will also cover how to run ipyrad on a cluster and how to do so efficiently.
Overview of assembly steps
Very roughly speaking, ipyrad exists to transform raw data coming off the sequencing instrument into output files that you can use for downstream analysis.
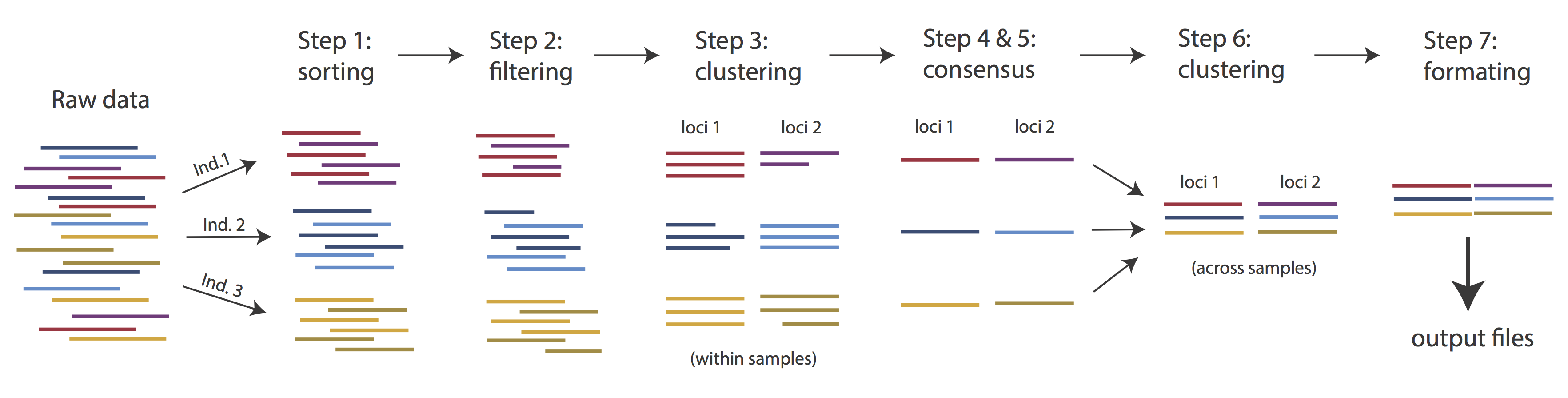
The basic steps of this process are as follows:
- Step 1 - Demultiplex/Load raw data
- Step 2 - Trim and quality control
- Step 3 - Cluster or reference-map within samples
- Step 4 - Calculate error rate and heterozygosity
- Step 5 - Call consensus sequences/alleles
- Step 6 - Cluster across samples
- Step 7 - Apply filters and write output formats
Detailed information about ipyrad, including instructions for installation and troubleshooting, can be found here.
Note: Assembling RADseq type sequence data requires a lot of different steps, and these steps generate a lot of intermediary files. Ipyrad organizes these files into directories, and it prepends the name of your assembly to each directory with data that belongs to it. One result of this is that you can have multiple assemblies of the same raw data with different parameter settings and you don’t have to manage all the files yourself! Another result is that you should not rename or move any of the directories inside your project directory, unless you know what you’re doing or you don’t mind if your assembly breaks. This is an important point when using pipelines, as file paths are usually included within the pipeline, and there are assumptions where the different files are.
Running ipyrad
Ipyrad needs a different environment, which was created from the yaml file in my input_files folder (if you haven’t done so yet, see instructions here). Load that environment first:
conda activate ipyrad
To better understand how to use ipyrad, let’s take a look at the help argument. We will use some of the ipyrad arguments in this tutorial (for example: -n, -p, -s, -c, -r). But, the complete list of optional arguments and their explanation is below.
ipyrad -h
usage: ipyrad [-h] [-v] [-r] [-f] [-q] [-d] [-n NEW] [-p PARAMS] [-s STEPS] [-b [BRANCH [BRANCH ...]]]
[-m [MERGE [MERGE ...]]] [-c cores] [-t threading] [--MPI] [--ipcluster [IPCLUSTER]]
[--download [DOWNLOAD [DOWNLOAD ...]]]
optional arguments:
-h, --help show this help message and exit
-v, --version show program's version number and exit
-r, --results show results summary for Assembly in params.txt and exit
-f, --force force overwrite of existing data
-q, --quiet do not print to stderror or stdout.
-d, --debug print lots more info to ipyrad_log.txt.
-n NEW create new file 'params-{new}.txt' in current directory
-p PARAMS path to params file for Assembly: params-{assembly_name}.txt
-s STEPS Set of assembly steps to run, e.g., -s 123
-b [BRANCH [BRANCH ...]]
create new branch of Assembly as params-{branch}.txt, and can be used to drop samples from
Assembly.
-m [MERGE [MERGE ...]]
merge multiple Assemblies into one joint Assembly, and can be used to merge Samples into one
Sample.
-c cores number of CPU cores to use (Default=0=All)
-t threading tune threading of multi-threaded binaries (Default=2)
--MPI connect to parallel CPUs across multiple nodes
--ipcluster [IPCLUSTER]
connect to running ipcluster, enter profile name or profile='default'
--download [DOWNLOAD [DOWNLOAD ...]]
download fastq files by accession (e.g., SRP or SRR)
* Example command-line usage:
ipyrad -n data ## create new file called params-data.txt
ipyrad -p params-data.txt -s 123 ## run only steps 1-3 of assembly.
ipyrad -p params-data.txt -s 3 -f ## run step 3, overwrite existing data.
* HPC parallelization across 32 cores
ipyrad -p params-data.txt -s 3 -c 32 --MPI
* Print results summary
ipyrad -p params-data.txt -r
* Branch/Merging Assemblies
ipyrad -p params-data.txt -b newdata
ipyrad -m newdata params-1.txt params-2.txt [params-3.txt, ...]
* Subsample taxa during branching
ipyrad -p params-data.txt -b newdata taxaKeepList.txt
* Download sequence data from SRA into directory 'sra-fastqs/'
ipyrad --download SRP021469 sra-fastqs/
* Documentation: http://ipyrad.readthedocs.io
Create a new parameters file
Ipyrad uses a text file to hold all the parameters for a given assembly. Start by creating a new parameters file with the -n flag. This flag
requires you to pass in a name for your assembly. In the example we use cheetah but the name can be anything at all. Once you start analysing your own data you might call your parameters file something more informative, including some details on the settings.
ipyrad -n cheetah
This will create a file in the current directory called params-cheetah.txt. The params file lists on each line one parameter followed by a ## mark, then the name of the parameter, and then a short description of its purpose. Lets take a look at it.
cat params-cheetah.txt
------- ipyrad params file (v.0.9.92)-------------------------------------------
cheetah ## [0] [assembly_name]: Assembly name. Used to name output directories for assembly steps
/home/uramakri/laurabertola/Tutorial_RADseq ## [1] [project_dir]: Project dir (made in curdir if not present)
## [2] [raw_fastq_path]: Location of raw non-demultiplexed fastq files
## [3] [barcodes_path]: Location of barcodes file
## [4] [sorted_fastq_path]: Location of demultiplexed/sorted fastq files
denovo ## [5] [assembly_method]: Assembly method (denovo, reference)
## [6] [reference_sequence]: Location of reference sequence file
rad ## [7] [datatype]: Datatype (see docs): rad, gbs, ddrad, etc.
TGCAG, ## [8] [restriction_overhang]: Restriction overhang (cut1,) or (cut1, cut2)
5 ## [9] [max_low_qual_bases]: Max low quality base calls (Q<20) in a read
33 ## [10] [phred_Qscore_offset]: phred Q score offset (33 is default and very standard)
6 ## [11] [mindepth_statistical]: Min depth for statistical base calling
6 ## [12] [mindepth_majrule]: Min depth for majority-rule base calling
10000 ## [13] [maxdepth]: Max cluster depth within samples
0.85 ## [14] [clust_threshold]: Clustering threshold for de novo assembly
0 ## [15] [max_barcode_mismatch]: Max number of allowable mismatches in barcodes
0 ## [16] [filter_adapters]: Filter for adapters/primers (1 or 2=stricter)
35 ## [17] [filter_min_trim_len]: Min length of reads after adapter trim
2 ## [18] [max_alleles_consens]: Max alleles per site in consensus sequences
0.05 ## [19] [max_Ns_consens]: Max N's (uncalled bases) in consensus (R1, R2)
0.05 ## [20] [max_Hs_consens]: Max Hs (heterozygotes) in consensus (R1, R2)
4 ## [21] [min_samples_locus]: Min # samples per locus for output
0.2 ## [22] [max_SNPs_locus]: Max # SNPs per locus (R1, R2)
8 ## [23] [max_Indels_locus]: Max # of indels per locus (R1, R2)
0.5 ## [24] [max_shared_Hs_locus]: Max # heterozygous sites per locus
0, 0, 0, 0 ## [25] [trim_reads]: Trim raw read edges (R1>, <R1, R2>, <R2) (see docs)
0, 0, 0, 0 ## [26] [trim_loci]: Trim locus edges (see docs) (R1>, <R1, R2>, <R2)
p, s, l ## [27] [output_formats]: Output formats (see docs)
## [28] [pop_assign_file]: Path to population assignment file
## [29] [reference_as_filter]: Reads mapped to this reference are removed in step 3
In general the defaults are sensible, and we won’t mess with them for now, but there are a few parameters we must check and update:
- The path to the raw data
- The dataype
- The restriction overhang sequence(s)
Because we’re looking at population-level data, we suggest to increase the clustering threshold [14] [clust_threshold]. You can also change [27] [output_formats]. When you put *, ipyrad will automatically save your output in all available formats, see the manual.
You can edit your params file with a text editor, for example nano. Type the following:
nano params-cheetah.txt
Now you can move around with the arrow keys and adjust the content of the file. We need to specify where the raw data files are located, the type of data we are using (.e.g., ‘gbs’, ‘rad’, ‘ddrad’, ‘pairddrad), and which enzyme cut site overhangs are expected to be present on the reads. Change the following lines in your params files to look like this:
./input_files/*.fastq.gz ## [4] [sorted_fastq_path]: Location of demultiplexed/sorted fastq files
rad ## [7] [datatype]: Datatype (see docs): rad, gbs, ddrad, etc.
CATGC ## [8] [restriction_overhang]: Restriction overhang (cut1,) or (cut1, cut2)
0.9 ## [14] [clust_threshold]: Clustering threshold for de novo assembly
* ## [27] [output_formats]: Output formats (see docs)
You exit the text editor with CTRL + X (note it lists the most common commands at the bottom of the screen). It will then ask you if you want to save the changes, so you’ll have to say Y.
Once we start running the analysis ipyrad will create several new directories to hold the output of each step for this assembly. By default the new directories are created in the project_dir directory and use the prefix specified by the assembly_name parameter. For this example assembly all the intermediate directories will be of the form: /Tutorial_RADseq/cheetah_*.
Step 1: Loading/Demultiplexing the raw data
Sometimes, you’ll receive your data as a huge pile of reads, and you’ll need to split it up and assign each read to the sample it came from. This is called demultiplexing and is done by unique barcodes which allow you to recognize individual samples. In that case, you’ll have to provide a path to the raw non-demultiplexed fastq files [2] and the path to the barcode file [3] in your params file. In our case, the samples are already demultiplexed and we have 1 file per sample. The path to these files is indicated in [4] in the params file. Even though we do not need to demultiplex our data here, we still need to run this step to import the data into ipyrad.
Note on step 1: If we would have data which need demultiplexing, Step 1 will create a new folder, called
cheetah_fastqs. Because our data are already demultiplexed, this folder will not be created.
Now lets run step 1!
ipyrad -p params-cheetah.txt -s 1 -c 4
-------------------------------------------------------------
ipyrad [v.0.9.92]
Interactive assembly and analysis of RAD-seq data
-------------------------------------------------------------
Parallel connection | osboxes: 4 cores
Step 1: Loading sorted fastq data to Samples
[####################] 100% 0:00:13 | loading reads
24 fastq files loaded to 24 Samples.
Parallel connection closed.
We use the following flags:
-p the params file we wish to use
-s the step to run
-c run on 4 cores
For those who are interested in what goes on under the hood:
Any time ipyrad is invoked it performs a few housekeeping operations:
- Load the assembly object - Since this is our first time running any steps we need to initialize our assembly.
- Start the parallel cluster - ipyrad uses a parallelization library called ipyparallel. Every time we start a step we fire up the parallel clients. This makes your assemblies go smokin’ fast.
- Do the work - Actually perform the work of the requested step(s) (in this case demultiplexing reads to samples).
- Save, clean up, and exit - Save the state of the assembly, and spin down the ipyparallel cluster.
As a convenience ipyrad internally tracks the state of all your steps in your current assembly, so at any time you can ask for results by invoking the -r flag. We also use the -p argument to tell it which params file (i.e., which assembly) we want it to print stats for.
ipyrad -p params-cheetah.txt -r
loading Assembly: cheetah
from saved path: ~/Tutorial_RADseq/cheetah.json
Summary stats of Assembly cheetah
------------------------------------------------
state reads_raw
SRR19760910 1 125000
SRR19760912 1 125000
SRR19760918 1 125000
SRR19760920 1 125000
SRR19760921 1 125000
SRR19760924 1 125000
SRR19760927 1 125000
SRR19760928 1 125000
SRR19760942 1 125000
SRR19760946 1 125000
SRR19760947 1 125000
SRR19760949 1 125000
SRR19760950 1 125000
SRR19760951 1 125000
SRR19760953 1 120739
SRR19760954 1 125000
SRR19760955 1 125000
SRR19760956 1 125000
SRR19760957 1 125000
SRR19760958 1 125000
SRR19760959 1 125000
SRR19760960 1 125000
SRR19760961 1 125000
SRR19760962 1 125000
Full stats files
------------------------------------------------
step 1: ./cheetah_s1_demultiplex_stats.txt
step 2: None
step 3: None
step 4: None
step 5: None
step 6: None
step 7: None
If you want to get even more info, ipyrad tracks all kinds of wacky stats and saves them to a file inside the directories it creates for each step.
Step 2: Filter reads
This step filters reads based on quality scores and maximum number of uncalled bases, and can be used to detect Illumina adapters in your reads, which is sometimes a problem under a couple different library prep scenarios. We know the our data have an excess of low-quality bases toward the 5’ end (remember the FastQC results!), so lets use this opportunity to trim off some of those low quality regions. To account for this we will trim reads to 100bp, removing the last 10bp of our 110bp reads.
Edit your params file again with nano and change the following parameter setting:
0, 100, 0, 0 ## [25] [trim_reads]: Trim raw read edges (R1>, <R1, R2>, <R2) (see docs)
Let’s now run step 2:
ipyrad -p params-cheetah.txt -s 2 -c 4
loading Assembly: cheetah
from saved path: ~/Tutorial_RADseq/cheetah.json
-------------------------------------------------------------
ipyrad [v.0.9.92]
Interactive assembly and analysis of RAD-seq data
-------------------------------------------------------------
Parallel connection | osboxes: 4 cores
Step 2: Filtering and trimming reads
[####################] 100% 0:01:29 | processing reads
Parallel connection closed.
The filtered files are written to a new directory called cheetah_edits. Again, you can look at the results from this step and some handy stats tracked for this assembly.
cat cheetah_edits/s2_rawedit_stats.txt
reads_raw trim_adapter_bp_read1 trim_adapter_bp_read2 trim_quality_bp_read1 trim_quality_bp_read2 reads_filtered_by_Ns reads_filtered_by_minlen reads_passed_filter
reads_raw trim_adapter_bp_read1 trim_quality_bp_read1 reads_filtered_by_Ns reads_filtered_by_minlen reads_passed_filter
SRR19760910 125000 3907 151184 59 266 124675
SRR19760912 125000 4316 232833 65 631 124304
SRR19760918 125000 4029 44448 3 37 124960
SRR19760920 125000 3981 46216 5 47 124948
SRR19760921 125000 4140 45663 14 31 124955
SRR19760924 125000 4355 42285 9 47 124944
SRR19760927 125000 4120 51549 11 52 124937
SRR19760928 125000 4068 44598 8 46 124946
SRR19760942 125000 4210 48041 12 250 124738
SRR19760946 125000 4538 39388 9 390 124601
SRR19760947 125000 4335 44185 14 39 124947
SRR19760949 125000 4383 187211 55 313 124632
SRR19760950 125000 3757 184015 52 393 124555
SRR19760951 125000 3956 261547 62 677 124261
SRR19760953 120739 4097 119160 49 268 120422
SRR19760954 125000 4176 238676 56 639 124305
SRR19760955 125000 3783 241923 56 584 124360
SRR19760956 125000 4290 178120 63 379 124558
SRR19760957 125000 4068 191626 49 410 124541
SRR19760958 125000 4290 177849 65 354 124581
SRR19760959 125000 4027 221129 74 523 124403
SRR19760960 125000 4127 171203 60 316 124624
SRR19760961 125000 3976 254342 46 640 124314
SRR19760962 125000 3948 220764 52 530 124418
ipyrad -p params-cheetah.txt -r
You might also take a closer look at the filtered reads:
zcat cheetah_edits/SRR19760910.trimmed_R1_.fastq.gz | head -n 12
@SRR19760910.1 1 length=110
CATGCACGTGCAGCATATAAGAAGGATGTTTGTCATGCATTATCTTATTTGATGTTTACGGAAGCCCCATGGTTATCCCCATTTTAGGGATGAAGAAACG
+
BFFFFFF<FBFFFFFF<FB//////<<FFFBFF//<FFFFFFBF/FBFFFFFFFFFFFFBB<F/BFFFFFFFFBFF/<<</BFBBFF/<FF<FF<7FFFF
@SRR19760910.2 2 length=110
CATGCAACTCTTGGTCTCGGGGTCTTGAGTTCGAGCCCCACGTTGGATTAGAGATTACTTAAATAAATAAAGTTCAAAAGTTTTAGAATGTTATCATTTT
+
FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF
@SRR19760910.3 3 length=110
CATGCCATTTCCCATGGGCAAGGATCTCAGGCTGTGCTCATTCCCAAGGACAAGACCAAGCCAATTCCCAATCCCCATATTTAAGGAGCTGCTTCCTGGG
+
FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF<FFFFFFFFFFFFFFFBFFFFFFFFFFFFFFFFFFFFFFFFFFBFFFFFFFFFBFFFFFFFFFFFFF
This is actually really cool, because we can already see the results of our applied parameters. All reads have been trimmed to 100bp.
Step 3: denovo clustering within-samples
For a de novo assembly, step 3 de-replicates and then clusters reads within each sample by the set clustering threshold and then writes the clusters to new files in a directory called cheetah_clust_0.9. Intuitively, we are trying to identify all the reads that map to the same locus within each sample. You may remember the default value is 0.85, but we have increased if to 0.9 in our params file. This value dictates the percentage of sequence similarity that reads must have in order to be considered reads at the same locus.
Note: The true name of this output directory will be dictated by the value you set for the
clust_thresholdparameter in the params file.
You’ll more than likely want to experiment with this value, but 0.9 is a reasonable default for population genetic-scale data, balancing over-splitting of loci vs over-lumping. Don’t mess with this until you feel comfortable with the overall workflow, and also until you’ve learned about branching assemblies.
It’s also possible to incorporate information from a reference genome to improve clustering at this step, if such a resources is available for your organism (or one that is relatively closely related). We will not cover reference based assemblies in this workshop, but you can refer to the ipyrad documentation for more information.
Note on performance: Steps 3 and 6 generally take considerably longer than any of the steps, due to the resource intensive clustering and alignment phases. These can take on the order of 10-100x as long as the next longest running step. This depends heavily on the number of samples in your dataset, the number of cores, the length(s) of your reads, and the “messiness” of your data.
Now lets run step 3:
ipyrad -p params-cheetah.txt -s 3 -c 4
loading Assembly: cheetah
from saved path: ~/Tutorial_RADseq/cheetah.json
-------------------------------------------------------------
ipyrad [v.0.9.92]
Interactive assembly and analysis of RAD-seq data
-------------------------------------------------------------
Parallel connection | osboxes: 4 cores
Step 3: Clustering/Mapping reads within samples
[####################] 100% 0:00:13 | dereplicating
[####################] 100% 0:09:46 | clustering/mapping
[####################] 100% 0:00:00 | building clusters
[####################] 100% 0:00:00 | chunking clusters
[####################] 100% 0:16:25 | aligning clusters
[####################] 100% 0:00:59 | concat clusters
[####################] 100% 0:00:05 | calc cluster stats
Parallel connection closed.
In-depth operations of step 3:
- dereplicating - Merge all identical reads
- clustering - Find reads matching by sequence similarity threshold
- building clusters - Group similar reads into clusters
- chunking clusters - Subsample cluster files to improve performance of alignment step
- aligning clusters - Align all clusters
- concat clusters - Gather chunked clusters into one full file of aligned clusters
- calc cluster stats - Just as it says.
Again we can examine the results. The stats output tells you how many clusters were found (‘clusters_total’), and the number of clusters that pass the mindepth thresholds (‘clusters_hidepth’). We’ll go into more detail about mindepth settings in some of the advanced tutorials.
ipyrad -p params-cheetah.txt -r
Summary stats of Assembly cheetah
------------------------------------------------
state reads_raw reads_passed_filter clusters_total clusters_hidepth
SRR19760910 3 125000 124675 36092 6433
SRR19760912 3 125000 124304 38568 6103
SRR19760918 3 125000 124960 34586 6989
SRR19760920 3 125000 124948 34759 6851
SRR19760921 3 125000 124955 34423 7003
SRR19760924 3 125000 124944 35112 6793
SRR19760927 3 125000 124937 34872 6941
SRR19760928 3 125000 124946 35102 6860
SRR19760942 3 125000 124738 36514 6629
SRR19760946 3 125000 124601 40697 6135
SRR19760947 3 125000 124947 37190 6262
SRR19760949 3 125000 124632 37495 6179
SRR19760950 3 125000 124555 33231 6092
SRR19760951 3 125000 124261 33844 6744
SRR19760953 3 120739 120422 33549 6216
SRR19760954 3 125000 124305 35873 6326
SRR19760955 3 125000 124360 33113 6186
SRR19760956 3 125000 124558 39780 5953
SRR19760957 3 125000 124541 37946 6190
SRR19760958 3 125000 124581 38539 6043
SRR19760959 3 125000 124403 38225 6172
SRR19760960 3 125000 124624 38138 6238
SRR19760961 3 125000 124314 36710 6333
SRR19760962 3 125000 124418 33510 7085
Full stats files
------------------------------------------------
step 1: ./cheetah_s1_demultiplex_stats.txt
step 2: ./cheetah_edits/s2_rawedit_stats.txt
step 3: ./cheetah_clust_0.9/s3_cluster_stats.txt
step 4: None
step 5: None
step 6: None
step 7: None
Again, the final output of step 3 is dereplicated, clustered files for each sample in ./cheetah_clust_0.9/. You can get a feel for what
this looks like by examining a portion of one of the files.
zcat cheetah_clust_0.9/SRR19760910.clustS.gz | head -n 100
You’ll see something similar to what is printed below:
0121ac19c8acb83e5d426007a2424b65;size=18;*
TGCAGTTGGGATGGCGATGCCGTACATTGGCGCATCCAGCCTCGGTCATTGTCGGAGATCTCACCTTTCAACGGTnnnnTGAATGGTCGCGACCCCCAACCACAATCGGCTTTGCCAAGGCAAGGCTAGAGACGTGCTAAAAAAACTCGCTCCG
521031ed2eeb3fb8f93fd3e8fdf05a5f;size=1;+
TGCAGTTGGGATGGCGATGCCGTACATTGGCGCATCCAGCCTCGGTCATTGTCGGAGATCTCACCTTTCAACGGTnnnnTGAATGGTCGCGACCCCCAACCACAATCGGCTTTGCCAAGGCAAGGCTAGAGAAGTGCTAAAAAAACTCGCTCCG
//
//
014947fbb43ef09f5388bbd6451bdca0;size=12;*
TGCAGGACTGCGAATGACGGTGGCTAGTACTCGAGGAAGGGTCGCACCGCAGTAAGCTAATCTGACCCTCTGGAGnnnnACCAGTGGTGGGTAAACACCTCCGATTAAGTATAACGCTACGTGAAGCTAAACGGCACCTATCACATAGACCCCG
072588460dac78e9da44b08f53680da7;size=8;+
TGCAGGTCTGCGAATGACGGTGGCTAGTACTCGAGGAAGGGTCGCACCGCAGTAAGCTAATCTGACCCTCTGGAGnnnnACCAGTGGTGGGTAAACACCTCCGATTAAGTATAACGCTACGTGAAGCTAAACGGCACCTATCACATAGACCCCG
fce2e729af9ea5468bafbef742761a4b;size=1;+
TGCAGGACTGCGAATGACGGTGGCTAGTACTCGAGGAAGGGTCGCACCGCAGCAAGCTAATCTGACCCTCTGGAGnnnnACCAGTGGTGGGTAAACACCTCCGATTAAGTATAACGCTACGTGAAGCTAAACGGCACCTATCACATAGACCCCG
24d23e93688f17ab0252fe21f21ce3a7;size=1;+
TGCAGGTCTGCGAATGACGGTGGCTAGTACTCGAGGAAGGGTCGCACCGCAGAAAGCTAATCTGACCCTCTGGAGnnnnACCAGTGGTGGGTAAACACCTCCGATTAAGTATAACGCTACGTGAAGCTAAACGGCACCTATCACATAGACCCCG
ef2c0a897eb5976c40f042a9c3f3a8ba;size=1;+
TGCAGGTCTGCGAATGACGGTGGCTAGTACTCGAGGAAGGGTCGCACCGCAGTAAGCTAATCTGACCCTCTGGAGnnnnACCAGTGGTGGGTAAACACCTCCGATTAAGTATAACGCTACGTGAAGCTAAACGGCACCTATCACATCGACCCCG
//
//
Reads that are sufficiently similar (based on the above sequence similarity threshold) are grouped together in clusters separated by “//”. The first cluster above is probably homozygous with some sequencing error. The second cluster is probably heterozygous with some sequencing error. We don’t want to go through and ‘decide’ by ourselves for each cluster, so thankfully, untangling this mess is what steps 4 & 5 are all about.
Step 4: Joint estimation of heterozygosity and error rate
In Step 3 reads that are sufficiently similar (based on the specified sequence similarity threshold) are grouped together in clusters separated by “//”. We examined the head of one of the sample cluster files at the end of the last exercise, but here we’ve cherry picked a couple clusters with more pronounced features.
Here’s a nice homozygous cluster, with probably one read with sequencing error:
0082e23d9badff5470eeb45ac0fdd2bd;size=5;*
TGCATGTAGTGAAGTCCGCTGTGTACTTGCGAGAGAATGAGTAGTCCTTCATGCA
a2c441646bb25089cd933119f13fb687;size=1;+
TGCATGTAGTGAAGTCCGCTGTGTACTTGCGAGAGAATGAGCAGTCCTTCATGCA
Here’s a probable heterozygote, or perhaps repetitive element – a little bit messier (note the indels):
0091f3b72bfc97c4705b4485c2208bdb;size=3;*
TGCATACAC----GCACACA----GTAGTAGTACTACTTTTTGTTAACTGCAGCATGCA
9c57902b4d8e22d0cda3b93f1b361e78;size=3;-
TGCATACAC----ACACACAACCAGTAGTAGTATTACTTTTTGTTAACTGCAGCATGCA
d48b3c7b5a0f1840f54f6c7808ca726e;size=1;+
TGCATACAC----ACAAACAACCAGTTGTAGTACTACTTTTTGTTAACTGCAGCATGAA
fac0c64aeb8afaa5dfecd5254b81b3c0;size=1;+
TGCATACAC----GCACACAACCAGTAGTAGTACTACTTTTTGTTAACTGCAGCATGTA
f31cbca6df64e7b9cb4142f57e607a88;size=1;-
TGCATGCACACACGCACGCAACCAGTAGTTGTACTACTTTTTGTTAACTGCAGCATGCA
935063406d92c8c995d313b3b22c6484;size=1;-
TGCATGCATACACGCCCACAACCAGTAGTAGTACAACTTTATGTTAACTGCAGCATGCA
d25fcc78f14544bcb42629ed2403ce74;size=1;+
TGCATACAC----GCACACAACCAGTAGTAGTACTACTTTTTGTTAATTGCAGCATGCA
Here’s a nasty one!
008a116c7a22d6af3541f87b36a8d895;size=3;*
TGCATTCCTATGGGAATCATGAAGGGGCTTCTCTCTCCCTCA-TTTTTAAAGCGACCCTTTCCAAACTTGGTACAT----
a7bde31f2034d2e544400c62b1d3cbd5;size=2;+
TGCATTCCTATGGGAAACATGAAGGGACTTCTCTCTCCCTCG-TTTTTAAAGTGACTCTGTCCAAACTTGGTACAT----
107e1390e1ac8564619a278fdae3f009;size=2;+
TGCATTCCTATGGGAAACATGAAGGGGGTTCTCTCTCCCTCG-ATTTTAAAGCGACCCTGTCCAAACTTGGTACAT----
8f870175fb30eed3027b7aec436e93e6;size=2;+
TGCATTCCTATGGGAATCATGGAAGGGCTTCTCTCTCCCTCA-TTTTTAAAGCAACCCTGACCAAAGTTGGTACAT----
445157bc1e7540734bf963eb8629d827;size=2;+
TGCATTCCTACGGGAATCATGGAGGGGCTTCTCTCTCCCTCG-TTTTTAAAGCGACCCTGACCAAACTTGGTACAT----
9ddd2d8b6fb52157f17648682d09afda;size=1;+
TGCATTCCTATGAGAAACATGATGGGGCTTCTCTTTCCCTCATTTTTT--AGTTAGCCTTACCAAAGTTGGTACATT---
fc86d48758313be18587d6f185e5c943;size=1;+
TGCATTCCTGTGGGAAACATGAAGGGGCTTCTCTCTCCATCA-TTTTTAAAGCGACCCTGATCAAATTTGGTACAT----
243a5acbee6cd9cd223252a8bb65667e;size=1;+
TGCATTCCTATGGGAAACATGAAAGGGTTTCTCTCTCCCTCG-TTTTAAAAGCGACCCTGTCCAAACATGGTACAT----
55e50e131ec21fce8021f22de49bb7be;size=1;+
TGCATTCCAATGGGAAACATGAAAGGGCTTCTCTCTCCCTCG-TTTTTAAAGCGACCCTGTCCAAACTTGGTACAT----
For this final cluster it’s really hard to call by eye, that’s why we make the computer do it!
In this step we jointly estimate sequencing error rate and heterozygosity to help us figure out which reads are “real” and which include sequencing error. We need to know which reads are “real” because in diploid organisms there are a maximum of 2 alleles at any given locus. If we look at the raw data and there are 20 different “alleles”, and 2 of them are very high frequency, and the rest are singletons then this gives us evidence that the 2 high frequency alleles are the good reads and the rest are probably junk. This step is pretty straightforward, and pretty fast. Run it like this:
ipyrad -p params-cheetah.txt -s 4 -c 4
loading Assembly: cheetah
from saved path: ~/Tutorial_RADseq/cheetah.json
-------------------------------------------------------------
ipyrad [v.0.9.92]
Interactive assembly and analysis of RAD-seq data
-------------------------------------------------------------
Parallel connection | osboxes: 4 cores
Step 4: Joint estimation of error rate and heterozygosity
[####################] 100% 0:01:46 | inferring [H, E]
Parallel connection closed.
In terms of results, there isn’t as much to look at as in previous steps, though you can invoke the -r flag to see the estimated heterozygosity and error rate per sample.
ipyrad -p params-cheetah.txt -r
Summary stats of Assembly cheetah
------------------------------------------------
state reads_raw reads_passed_filter clusters_total clusters_hidepth hetero_est error_est
SRR19760910 4 125000 124675 36092 6433 0.001586 0.001939
SRR19760912 4 125000 124304 38568 6103 0.001574 0.001457
SRR19760918 4 125000 124960 34586 6989 0.001941 0.001793
SRR19760920 4 125000 124948 34759 6851 0.001958 0.001718
SRR19760921 4 125000 124955 34423 7003 0.001757 0.001781
SRR19760924 4 125000 124944 35112 6793 0.001780 0.001774
SRR19760927 4 125000 124937 34872 6941 0.001694 0.001987
SRR19760928 4 125000 124946 35102 6860 0.001663 0.001771
SRR19760942 4 125000 124738 36514 6629 0.001523 0.001638
SRR19760946 4 125000 124601 40697 6135 0.001910 0.001771
SRR19760947 4 125000 124947 37190 6262 0.002124 0.001934
SRR19760949 4 125000 124632 37495 6179 0.002958 0.001242
SRR19760950 4 125000 124555 33231 6092 0.000969 0.001323
SRR19760951 4 125000 124261 33844 6744 0.001339 0.001439
SRR19760953 4 120739 120422 33549 6216 0.001850 0.002019
SRR19760954 4 125000 124305 35873 6326 0.001581 0.001559
SRR19760955 4 125000 124360 33113 6186 0.001643 0.001609
SRR19760956 4 125000 124558 39780 5953 0.001578 0.001316
SRR19760957 4 125000 124541 37946 6190 0.001664 0.001254
SRR19760958 4 125000 124581 38539 6043 0.001611 0.001330
SRR19760959 4 125000 124403 38225 6172 0.001353 0.001430
SRR19760960 4 125000 124624 38138 6238 0.001306 0.001118
SRR19760961 4 125000 124314 36710 6333 0.001388 0.001479
SRR19760962 4 125000 124418 33510 7085 0.001505 0.001343
Full stats files
------------------------------------------------
step 1: ./cheetah_s1_demultiplex_stats.txt
step 2: ./cheetah_edits/s2_rawedit_stats.txt
step 3: ./cheetah_clust_0.9/s3_cluster_stats.txt
step 4: ./cheetah_clust_0.9/s4_joint_estimate.txt
step 5: None
step 6: None
step 7: None
Illumina error rates are on the order of 0.1% per base, so your error rates will ideally be in this neighborhood. Also, under normal conditions error rate will be much, much lower than heterozygosity (on the order of 10x lower). If the error rate is »0.1% then you might be using too permissive a clustering threshold. Just a thought.
Step 5: Consensus base calls
Step 5 uses the inferred error rate and heterozygosity per sample to call the consensus of sequences within each cluster. Here we are identifying what we believe to be the real haplotypes at each locus within each sample.
ipyrad -p params-cheetah.txt -s 5 -c 4
loading Assembly: cheetah
from saved path: ~/Tutorial_RADseq/cheetah.json
-------------------------------------------------------------
ipyrad [v.0.9.92]
Interactive assembly and analysis of RAD-seq data
-------------------------------------------------------------
Parallel connection | osboxes: 4 cores
Step 5: Consensus base/allele calling
Mean error [0.00158 sd=0.00026]
Mean hetero [0.00168 sd=0.00037]
[####################] 100% 0:00:04 | calculating depths
[####################] 100% 0:00:05 | chunking clusters
[####################] 100% 0:10:05 | consens calling
[####################] 100% 0:00:19 | indexing alleles
Parallel connection closed.
In-depth operations of step 5:
- calculating depths - A simple refinement of the H/E estimates
- chunking clusters - Again, breaking big files into smaller chunks to aid parallelization
- consensus calling - Actually perform the consensus sequence calling
- indexing alleles - Cleaning up and re-joining chunked data
ipyrad -p params-cheetah.txt -r
loading Assembly: cheetah
from saved path: ~/Tutorial_RADseq/cheetah.json
Summary stats of Assembly cheetah
------------------------------------------------
state reads_raw reads_passed_filter clusters_total clusters_hidepth hetero_est error_est reads_consens
SRR19760910 5 125000 124675 36092 6433 0.001586 0.001939 6355
SRR19760912 5 125000 124304 38568 6103 0.001574 0.001457 6036
SRR19760918 5 125000 124960 34586 6989 0.001941 0.001793 6905
SRR19760920 5 125000 124948 34759 6851 0.001958 0.001718 6766
SRR19760921 5 125000 124955 34423 7003 0.001757 0.001781 6930
SRR19760924 5 125000 124944 35112 6793 0.001780 0.001774 6724
SRR19760927 5 125000 124937 34872 6941 0.001694 0.001987 6867
SRR19760928 5 125000 124946 35102 6860 0.001663 0.001771 6793
SRR19760942 5 125000 124738 36514 6629 0.001523 0.001638 6566
SRR19760946 5 125000 124601 40697 6135 0.001910 0.001771 6072
SRR19760947 5 125000 124947 37190 6262 0.002124 0.001934 6182
SRR19760949 5 125000 124632 37495 6179 0.002958 0.001242 6090
SRR19760950 5 125000 124555 33231 6092 0.000969 0.001323 6043
SRR19760951 5 125000 124261 33844 6744 0.001339 0.001439 6677
SRR19760953 5 120739 120422 33549 6216 0.001850 0.002019 6142
SRR19760954 5 125000 124305 35873 6326 0.001581 0.001559 6255
SRR19760955 5 125000 124360 33113 6186 0.001643 0.001609 6102
SRR19760956 5 125000 124558 39780 5953 0.001578 0.001316 5897
SRR19760957 5 125000 124541 37946 6190 0.001664 0.001254 6124
SRR19760958 5 125000 124581 38539 6043 0.001611 0.001330 5976
SRR19760959 5 125000 124403 38225 6172 0.001353 0.001430 6117
SRR19760960 5 125000 124624 38138 6238 0.001306 0.001118 6178
SRR19760961 5 125000 124314 36710 6333 0.001388 0.001479 6269
SRR19760962 5 125000 124418 33510 7085 0.001505 0.001343 7018
Full stats files
------------------------------------------------
step 1: ./cheetah_s1_demultiplex_stats.txt
step 2: ./cheetah_edits/s2_rawedit_stats.txt
step 3: ./cheetah_clust_0.9/s3_cluster_stats.txt
step 4: ./cheetah_clust_0.9/s4_joint_estimate.txt
step 5: ./cheetah_consens/s5_consens_stats.txt
step 6: None
step 7: None
And here the important information is the number of reads_consens. This is the number of retained reads within each sample that we’ll send on to the next step. Retained reads must pass filters on read depth tolerance (both mindepth_majrule and maxdepth), maximum number of uncalled bases (max_Ns_consens) and maximum number of heterozygous sites (max_Hs_consens) per consensus sequence.
Step 6: Cluster across samples
Step 6 clusters consensus sequences across samples. Now that we have good estimates for haplotypes within samples we can try to identify similar sequences at each locus among samples. We use the same clustering threshold as step 3 to identify sequences among samples that are probably sampled from the same locus, based on sequence similarity.
ipyrad -p params-cheetah.txt -s 6 -c 4
loading Assembly: cheetah
from saved path: ~/Tutorial_RADseq/cheetah.json
-------------------------------------------------------------
ipyrad [v.0.9.92]
Interactive assembly and analysis of RAD-seq data
-------------------------------------------------------------
Parallel connection | osboxes: 4 cores
Step 6: Clustering/Mapping across samples
[####################] 100% 0:00:03 | concatenating inputs
[####################] 100% 0:01:18 | clustering across
[####################] 100% 0:00:06 | building clusters
[####################] 100% 0:02:55 | aligning clusters
Parallel connection closed.
In-depth operations of step 6:
- concatenating inputs - Gathering all consensus files and preprocessing to improve performance
- clustering across - Cluster by similarity threshold across samples
- building clusters - Group similar reads into clusters
- aligning clusters - Align within each cluster
Since in general the stats for results of each step are sample based, the output of -r will only display what we had seen after step 5, so this is not that informative.
It might be more enlightening to consider the output of step 6 by examining the file that contains the reads clustered across samples:
cat cheetah_across/cheetah_clust_database.fa | head -n 27
>SRR19760910_0
CATGCATCTAAAATTATAGAGTACTCATGTTTTACAAAGGAATTCAGCCTGTTTTACAAACACATCAGGTGAGCATCACCCACCTTAGCTAATGTTATGG
>SRR19760912_0
CATGCATCTAAAATTATAGAGTACTCATGTTTTACAAAGGAATTCAGCCTGTTTTACAAACACATCAGGTGAGCATCACCCACCTTAGCTAATGTTATGG
>SRR19760919_1
CATGCATCTAAAATTATAGAGTACTCATGTTTTACAAAGGAATTCAGCCTGTTTTACAAACACATCAGGTGAGCATCACCCACCTTAGCTAATGTTATGG
>SRR19760947_0
CATGCATCTAAAATTATAGAGTACTCATGTTTTACAAAGGAATTCAGCCTGTTTTACAAACACATCAGGTGAGCATCACCCACCTTAGCTAATGTTATGG
>SRR19760951_1
CATGCATCTAAAATTATAGAGTACTCATGTTTTACAAAGGAATTCAGCCTGTTTTACAAACACATCAGGTGAGCATCACCCACCTTAGCTAATGTTATGG
>SRR19760954_5064
CATGCATCTAAAATTATAGAGTACTCATGTTTTACAAAGGAATTCAGCCTGTTTTACAAACACATCAGGTGAGCATCACCCACCTTAGCTAATGTTATGG
>SRR19760957_5406
CATGCATCTAAAATTATAGAGTACTCATGTTTTACAAAGGAATTCAGCCTGTTTTACAAACACATCAGGTGAGCATCACCCACCTTAGCTAATGTTATGG
>SRR19760961_5265
CATGCATCTAAAATTATAGAGTACTCATGTTTTACAAAGGAATTCAGCCTGTTTTACAAACACATCAGGTGAGCATCACCCACCTTAGCTAATGTTATGG
//
//
>SRR19760910_10
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
>SRR19760911_4305
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
>SRR19760913_9
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
>SRR19760920_12
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
>SRR19760921_14
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
>SRR19760951_11
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
>SRR19760952_11
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
>SRR19760957_12
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
>SRR19760958_8
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
>SRR19760962_9
CATGCTCTGCTCTGCAGCCTGCAGTCTTTATGTTTGCTCTATGTCATAAGAATTCTGGCATACTTGTTTCTGTGAAATACCTGTATTTAGAGAACAGACG
//
//
The final output of step 6 is a file in cheetah_across called cheetah_clust_database.fa. This file contains all aligned reads across all
samples. Executing the above command you’ll see all the reads that align at each locus. You’ll see the sample name of each read followed by the sequence of the read at that locus for that sample. If you wish to examine more loci you can increase the number of lines you want to view by increasing the value you pass to head in the above command (e.g. ... | head -n 300).
Step 7: Filter and write output files
The final step is to filter the data and write output files in many convenient file formats. First, we apply filters for maximum number of indels per locus, max heterozygosity per locus, max number of snps per locus, and minimum number of samples per locus. All these filters are configurable in the params file. You are encouraged to explore different settings, but the defaults are quite good and quite conservative.
To run step 7:
ipyrad -p params-cheetah.txt -s 7 -c 4
loading Assembly: cheetah
from saved path: ~/Tutorial_RADseq/cheetah.json
-------------------------------------------------------------
ipyrad [v.0.9.92]
Interactive assembly and analysis of RAD-seq data
-------------------------------------------------------------
Parallel connection | osboxes:: 4 cores
Step 7: Filtering and formatting output files
[####################] 100% 0:00:16 | applying filters
[####################] 100% 0:00:09 | building arrays
[####################] 100% 0:00:13 | writing conversions
[####################] 100% 0:00:28 | indexing vcf depths
[####################] 100% 0:00:05 | writing vcf output
Parallel connection closed.
In-depth operations of step 7:
- applying filters - Apply filters for max # indels, SNPs, & shared hets, and minimum # of samples per locus
- building arrays - Construct the final output data in hdf5 format
- writing conversions - Write out all designated output formats
Step 7 generates output files in the cheetah_outfiles directory. All the output formats specified by the output_formats parameter will be generated here. Let’s see what’s been created:
ls cheetah_outfiles/
cheetah.alleles cheetah.nex cheetah.snpsmap cheetah.usnps
cheetah.geno cheetah.phy cheetah_stats.txt cheetah.ustr
cheetah.gphocs cheetah.seqs.hdf5 cheetah.str cheetah.vcf
cheetah.loci cheetah.snps cheetah.treemix
cheetah.migrate cheetah.snps.hdf5 cheetah.ugeno
Ipyrad always creates the cheetah.loci file, as this is our internal format, as well as the cheetah_stats.txt file, which reports final statistics for the assembly (more below). The other files created fall in to 2 categories: files that contain the full sequence (i.e. the cheetah.phy and cheetah.seqs.hdf5 files) and files that contain only variable sites (i.e. the cheetah.snps and cheetah.snps.hdf5 files). The cheetah.snpsmap is a file which maps SNPs to loci, which is used downstream in the analysis toolkit for sampling unlinked SNPs. You’ll also see that there are some output files which have a ‘u’ in there format, e.g. .ustr and .ugeno. These files contain only one SNP per locus, while the normal .str and .geno file may contain SNPs from the same locus. For downstream analyses, it is often good to use data with a single SNP per locus to avoid linkage.
The most informative, human-readable file here is cheetah_stats.txt which gives extensive and detailed stats about the final assembly. A quick overview of the different sections of this file:
cat cheetah_outfiles/cheetah_stats.txt
## The number of loci caught by each filter.
## ipyrad API location: [assembly].stats_dfs.s7_filters
total_filters applied_order retained_loci
total_prefiltered_loci 0 0 22587
filtered_by_rm_duplicates 21 21 22566
filtered_by_max_indels 32 32 22534
filtered_by_max_SNPs 2 2 22532
filtered_by_max_shared_het 105 105 22427
filtered_by_min_sample 7275 7275 15152
total_filtered_loci 7435 7435 15152
This block indicates how filtering is impacting your final dataset. Each filter is applied in order from top to bottom, and the number of loci removed because of each filter is shown in the applied_order column. The total number of retained_loci after each filtering step is displayed in the final column. This is a good place for inspecting how your filtering thresholds are impacting your final dataset. For example, you might see that most loci are being filterd by min_sample_locus (a very common result), in which case you might reduce this threshold in your params file and re-run step 7 in order to retain more loci. You can use branching, so you can re-run part of the analysis, without overwriting the output you already generated.
The next block shows a simple summary of the number of loci retained for each sample in the final dataset. Pretty straightforward. If you have some samples that have very low sample_coverage here it might be good to remove them and re-run step 7. Also this can be done by using branching.
## The number of loci recovered for each Sample.
## ipyrad API location: [assembly].stats_dfs.s7_samples
sample_coverage
SRR19760910 5347
SRR19760912 5275
SRR19760918 5782
SRR19760920 5807
SRR19760921 5859
SRR19760924 5783
SRR19760927 5935
SRR19760928 5909
SRR19760942 5660
SRR19760946 5033
SRR19760947 4477
SRR19760949 3831
SRR19760950 4847
SRR19760951 5556
SRR19760953 5038
SRR19760954 4289
SRR19760955 4685
SRR19760956 4946
SRR19760957 5295
SRR19760958 4727
SRR19760959 5451
SRR19760960 5429
SRR19760961 5626
SRR19760962 5903
The next block is locus_coverage, which indicates the number of loci that contain exactly a given number of samples, and sum_coverage is just the running total of these in ascending order. So here, if it weren’t being filtered, locus coverage in the 1 column would indicate singletons (only one sample at this locus), and locus coverage in the 10 column indicates loci with full coverage (all samples have data at these loci).
Note: It’s important to notice that locus coverage below your
min_sample_locusparameter setting will all naturally equal 0, since by definition these are being removed.
## The number of loci for which N taxa have data.
## ipyrad API location: [assembly].stats_dfs.s7_loci
locus_coverage sum_coverage
1 0 0
2 0 0
3 0 0
4 2498 2498
5 2143 4641
6 1783 6424
7 1548 7972
8 1310 9282
9 1065 10347
10 927 11274
11 772 12046
12 664 12710
13 512 13222
14 470 13692
15 363 14055
16 293 14348
17 218 14566
18 200 14766
19 157 14923
20 102 15025
21 68 15093
22 38 15131
23 12 15143
24 9 15152
Whereas the previous block indicated samples per locus, below we are looking at SNPs per locus. In a similar fashion as above, these columns record the counts of loci containing given numbers of variable sites and parsimony informative sites (pis). The sum_* columns simply indicate the running total in ascending order.
Note: This block can be a little tricky because loci can end up getting double-counted. For example, a locus with 1 pis, and 2 autapomorphies will be counted once in the 3 row for
var, and once in the 1 row forpis. Apply care when interpreting these values.
The distribution of SNPs (var and pis) per locus.
## var = Number of loci with n variable sites (pis + autapomorphies)
## pis = Number of loci with n parsimony informative site (minor allele in >1 sample)
## ipyrad API location: [assembly].stats_dfs.s7_snps
## The "reference" sample is included if present unless 'exclude_reference=True'
var sum_var pis sum_pis
0 10775 0 13802 0
1 2793 2793 1163 1163
2 917 4627 118 1399
3 365 5722 39 1516
4 155 6342 10 1556
5 65 6667 8 1596
6 37 6889 3 1614
7 18 7015 5 1649
8 11 7103 2 1665
9 5 7148 1 1674
10 2 7168 0 1674
11 4 7212 0 1674
12 1 7224 1 1686
13 3 7263 0 1686
14 1 7277 0 1686
The next block displays statistics for each sample in the final dataset. Many of these stats will already be familiar, but this provides a nice compact view on how each sample is represented in the output. The one new stat here is loci_in_assembly, which indicates how many loci each sample has data for.
## Final Sample stats summary
state reads_raw reads_passed_filter clusters_total clusters_hidepth hetero_est error_est reads_consens loci_in_assembly
SRR19760910 7 125000 124675 36092 6433 0.001586 0.001939 6355 5347
SRR19760912 7 125000 124304 38568 6103 0.001574 0.001457 6036 5275
SRR19760918 7 125000 124960 34586 6989 0.001941 0.001793 6905 5782
SRR19760920 7 125000 124948 34759 6851 0.001958 0.001718 6766 5807
SRR19760921 7 125000 124955 34423 7003 0.001757 0.001781 6930 5859
SRR19760924 7 125000 124944 35112 6793 0.001780 0.001774 6724 5783
SRR19760927 7 125000 124937 34872 6941 0.001694 0.001987 6867 5935
SRR19760928 7 125000 124946 35102 6860 0.001663 0.001771 6793 5909
SRR19760942 7 125000 124738 36514 6629 0.001523 0.001638 6566 5660
SRR19760946 7 125000 124601 40697 6135 0.001910 0.001771 6072 5033
SRR19760947 7 125000 124947 37190 6262 0.002124 0.001934 6182 4477
SRR19760949 7 125000 124632 37495 6179 0.002958 0.001242 6090 3831
SRR19760950 7 125000 124555 33231 6092 0.000969 0.001323 6043 4847
SRR19760951 7 125000 124261 33844 6744 0.001339 0.001439 6677 5556
SRR19760953 7 120739 120422 33549 6216 0.001850 0.002019 6142 5038
SRR19760954 7 125000 124305 35873 6326 0.001581 0.001559 6255 4289
SRR19760955 7 125000 124360 33113 6186 0.001643 0.001609 6102 4685
SRR19760956 7 125000 124558 39780 5953 0.001578 0.001316 5897 4946
SRR19760957 7 125000 124541 37946 6190 0.001664 0.001254 6124 5295
SRR19760958 7 125000 124581 38539 6043 0.001611 0.001330 5976 4727
SRR19760959 7 125000 124403 38225 6172 0.001353 0.001430 6117 5451
SRR19760960 7 125000 124624 38138 6238 0.001306 0.001118 6178 5429
SRR19760961 7 125000 124314 36710 6333 0.001388 0.001479 6269 5626
SRR19760962 7 125000 124418 33510 7085 0.001505 0.001343 7018 5903
The final block displays some very brief, but informative, summaries of missingness in the assembly at both the sequence and the SNP level:
## Alignment matrix statistics:
snps matrix size: (24, 7277), 62.27% missing sites.
sequence matrix size: (24, 1571595), 65.34% missing sites.
Note: Sometimes you want to rerun a step that you’ve run before, and overwrite the results you already obtained. You can do that by adding the
-fflag, forcing ipyrad to overwrite already existing files. Remember that if you don’t want to overwrite existing data, you may want to use branching.
Congratulations! You’ve completed your first RAD-Seq assembly. Now you can try applying what you’ve learned to assemble your own real data. Please consult the ipyrad online documentation for details about many of the more powerful features of ipyrad, including reference sequence mapping, assembly branching, and the extensive analysis toolkit, which includes extensive downstream analysis tools for such things as clustering and population assignment, phylogenetic tree inference, quartet-based species tree inference, and much more.
There is also an exercise which explains branching in more detail here.
 ©Laura Bertola
©Laura Bertola